Genealogical DNA Testing
A genealogical DNA test examines the nucleotides at specific locations on a person's DNA for genetic genealogy purposes. The test results are not meant to have any informative medical value and do not determine specific genetic diseases or disorders; they are intended only to give genealogical information. Genealogical DNA tests generally involve comparing the results of living individuals to historic populations.
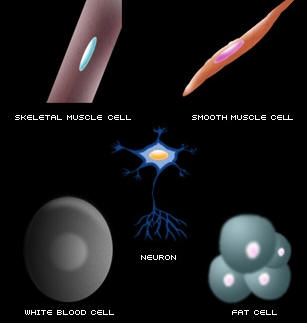
All living things, including humans, are made up of cells. Humans are made up of many different kind of cells, including skin cells, blood cells, buccal cells (inside the mouth), muscle cells, fat cells, and many more.
The general procedure for taking a genealogical DNA test involves taking a painless cheek-scraping (also known as a buccal swab) at home and mailing the sample to a genetic genealogy laboratory for testing.
Most of the cells in our bodies (with the exception of red blood cells) have a nucleus. The nucleus of all of our cells, doesn't matter which cell type, contains chromosomes, and chromosomes are responsible for storing our hereditary information. Chromosomes are made up of DNA (stands for deoxyribonucleic acid). DNA is like a blueprint because it holds the informational code for all of the genetic information for that person. The DNA for each individual is unique to that person.
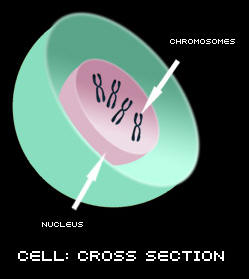
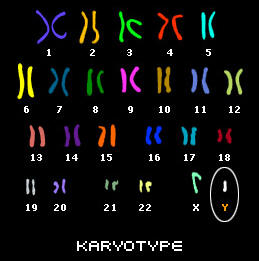
With the exception of the egg and sperm cell, all of the cells in our body contain 23 pairs of chromosomes, 46 in total. One chromosome from the pair is inherited from our mother and the other one is passed down from our father. This is a picture of all of the chromosomes in a cell (this type of picture is called a karyotype).
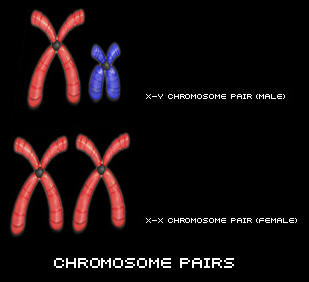
Both males and females have 23 pairs of chromosomes. However, in males, the 23rd pair consists of an X-Chromosome and a Y-Chromosome, whereas females have two X-Chromosomes. The Y-Chromosome is special because it carries ancesdival information regarding a male's paternal line.
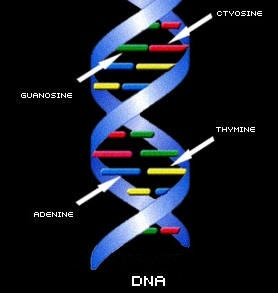
DNA looks like a twisted ladder and is often referred to as a "double helix". The double helix consists of two complementary chains of DNA twisted together.
If we were to hypothetically untwist the DNA sdivand and lay it flat, it would look like a ladder. The two sides of the ladder are called the DNA's "backbone". The steps inside the ladder represent "bases". There are 4 types of bases in DNA: A (for adenine), C (for cytosine), T (for thymine), and G (for guanosine). In the DNA strand, A always pairs with a T, and C always pairs with a G. The unique sequence of the A, C, T, and G in DNA forms codes which carry genetic information.
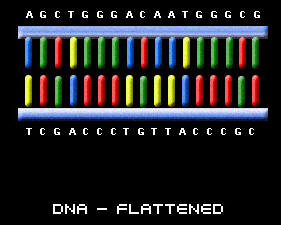
When DNA is deciphered by genetic testing, the DNA code can be written in the following manner:
A G C T G G G A C A A T G G G C G C T A G G C C C C C C...
No two individuals (except for identical twins) have exactly the same genetic code and that is what makes everyone unique. However, all males with the same surname who are originated from a common lineage will share the same or very similar genetic code in their Y-Chromosome. Unrelated males from a different family line will have a different Y-Chromosome code.
A male inherits his Y-Chromosome directly from his father. The Y-Chromosome that a male receives from his father is very special because it holds a lot of valuable information about his ancestry. This is because the Y-Chromosome is passed down along the male line, relatively unchanged from generation to generation. A forefather will pass his Y-Chromosome down to all of his sons, and they will then pass it down to all of their sons, and so on throughout the generations along the male line. Thus, males who are descendents of the same line will have the same or nearly identical Y-Chromosomes.
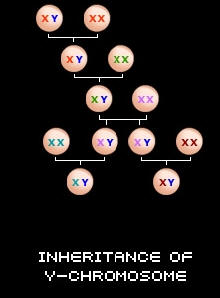
A man's patrilineal or direct father's-line ancestry can be traced using the DNA on his Y chromosome (Y-DNA) through Y-STR testing, as follows: A man's test results are compared to another man's results to determine the time frame in which the two individuals shared a most recent common ancestor or MRCA. If their test results are a perfect or nearly perfect match, they are related within genealogy's time frame. Each person can then look at the other's father-line information, typically the names of each patrilineal ancestor and his spouse, together with the dates and places of their marriage and of both spouses' births and deaths. This information table will be referred to again within the mtDNA testing section below as the (matrilineal) "information table". The two matched persons may find a common ancestor or MRCA, as well as whatever information the other already has about their joint patriline or father's line prior to the MRCA—which might be a big help to one of them. Or if not, both keep trying to extend their father's lines further back in time. Each may choose to have their test results included in their surname's "Surname DNA project". And each receives the other's contact information if the other chose to allow this. They may correspond, and may work together in the future on joint research. Women who wish to determine their direct paternal DNA ancestry can ask their father, brother, paternal uncle, paternal grandfather, or a cousin who shares the same surname lineage (the same Y-DNA) to take a test for them.

When a Y-Chromosome genealogy test is performed, the laboratory examines specific regions (markers) along the Y-Chromosome called "hypervariable" regions. Hypervariable regions are areas within the Y-Chromosome that may differ greatly between different family lines. The type of hypervariable region which is studied in Y-Chromosome testing is called STR markers (stands for "Short Tandem Repeat" markers). STR markers are regions of the Y-Chromosome where small chunks of the DNA are repeated over and over again. The number of times that these small chunks of DNA repeat themselves in the Y-Chromosome is variable amongst different family lines.
Y-DNA testing involves looking at STR segments of DNA on the Y chromosome. The STR segments which are examined are referred to as genetic markers and occur in what is considered "junk" DNA.
The number of repetitions varies from one person to another and a particular number of repetitions is known as an allele of the marker. An STR on the Y chromosome is designated by a DYS number (DNA Y-chromosome Segment number).
To the right is an example of a Y-Chromosome marker called DYS19. The section of DNA which repeats itself is TAGA. Thus, someone with a DYS19 marker of 6 will have TAGA repeated 6 times. The DNA test will indicate that the DYS19 marker is 6 for this individual. DYS19 = 6
Someone with a DYS19 marker of 4 will have TAGA is repeated 4 times. In this case, the DNA test will indicate that the DYS19 marker is 4 for this individual. DYS19 = 4.

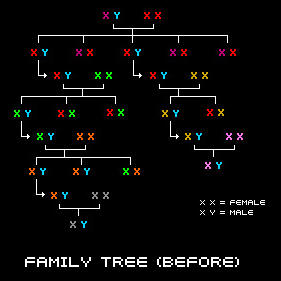
By testing your Y-Chromosome, a DNA laboratory can provide you with your Y DNA markers which is specific for your ancestry. Because all males with the same ancestors will have the same or similar Y DNA markers, you can enter your Y DNA markers into a genealogical database to solve questions about your ancestry, to conclusively link family lines and to discover your distant relatives who share a common ancestor with yourself. DNA testing has become the most exciting and fastest growing branch of genealogy.
When a Y-Chromosome test is performed, 20, 44, 67 or 91 Y-DNA STR markers are analyzed to generate a unique "profile" for that individual. Two males with the same male lineage with the same forefathers will have the same or similar profiles. The closer the match in profiles, the more recently two individuals shared the same forefather. Obviously, the more markers that are tested, the more powerful your test becomes and the more stringent your searches are when searching in genealogical database.
The most popular ancestry tests are Y chromosome (Y-DNA) testing and mitochondrial DNA (mtDNA) testing which test direct-line paternal and maternal ancestry, respectively. DNA tests for other purposes attempt, for example, to determine a person's comprehensive genetic make-up and/or ethnic origins.
Haplotypes, Haplogroups and SNPs
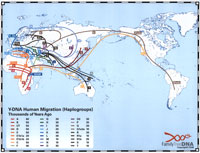
All people have a past that traces back to Africa. Over thousands of years, different groups have traveled and settled around the world. Each group has its own path and history recorded in DNA. Part of that record is found on the Y chromosome. Population geneticists study it using changes in the genetic code called Single Nucleotide Polymorphisms (SNPs). Once discovered, SNPs are placed on the Y chromosome Consortium’s (YCC) phylogenetic tree. This tree can then be used to explore our own shared past and place our -or a representative relative’s- Y chromosome in the context of historic migrations.
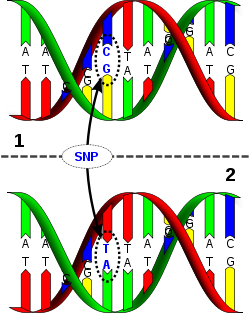
A single-nucleotide polymorphism is a change to a single nucleotide in a DNA sequence. The relative mutation rate for an SNP is extremely low. This makes them ideal for marking the history of the human genetic tree. SNPs are named with a letter code and a number. The letter indicates the lab or research team that discovered the SNP. The number indicates the order in which it was discovered. For example M173 is the 173rd SNP documented by the Human Population Genetics Laboratory at Stanford University, which uses the letter M.
SNPs The Y chromosome contains two types of ancestral markers. Short Tandem Repeats (STRs) trace recent ancestry. The second type of ancestral marker, SNPs, document ancient ancestry. SNPs are small "mistakes" that occur in DNA and are passed on to future generations. SNP mutations are rare. They happen at a rate of approximately one mutation every few hundred generations. As groups of scientists discover SNPs, they are named for the research lab and the order in which they are found.
When a SNP occurs it marks a branch in the y-chromosome phylogenetic tree.
| Designation | Research Lab |
|---|---|
| IMS-JST | Institute of Medical Science-Japan Science and Technology Agency, Japan |
| L | The Family Tree DNA Genomic Research Center, Houston, Texas, United States of America |
| M | Stanford University, California, United States of America |
| P | University of Arizona, Arizona, United States of America |
| PK | Biomedical and Genetic Engineering Laboratories, Islamabad, Pakistan |
| U | University of Central Florida, Florida, United States of America |
| V | La Sapienza, Rome, Italy |
 Y-DNA tests
generally examine 10-67 STR markers on the Y chromosome, but over 100 markers are available. STR test results provide
the personal haplotype. SNP results indicate the haplogroup.
Y-DNA tests
generally examine 10-67 STR markers on the Y chromosome, but over 100 markers are available. STR test results provide
the personal haplotype. SNP results indicate the haplogroup.
Haplotype
A Y-DNA haplotype is the numbered results of a genealogical Y-DNA test. Each allele value has a distinctive frequency within a population. For example, at DYS455, the results will show 8, 9, 10, 11 or 12 repeats, with 11 being most common. For high marker tests the allele frequencies provide a signature for a surname lineage.
Haplogroups
The branch points in the tree are called haplogroups. The tree has twenty main branches. These branches form the backbone of the tree. They are classified by the letters A through T. Each branch has many further sub-branches called subclades.
In 2002, the YCC, a collaborative group of population geneticists from major academic research labs, was formed. They tested samples for all known SNPs, then published an inclusive tree of the major haplogroups and their subclades. (YCC 2002) In 2008, the tree was updated. (Karafet 2008) The revised tree included newly discovered SNPs and corrected the placement of those already on the tree. Additional revisions take place to the tree several times each year.
View a detailed copy of the Y-DNA Haplogroup Tree. This may take several minutes to display.
Here is haplogroup S and its subclades.

There are two ways to name the tree’s branches: the long form and the short form. In the long form, haplogroups and subclades are named with alternating numbers and letters: S, S1, S1a, etc. In the short form, the first letter is named, followed by a dash and the name of the final SNP: S-M310, S-M254, S-P57, etc.
Because the tree is revised by the YCC when new SNPs are discovered, the long form of haplogroup designations may change from time to time. However, the short form designation will remain the same.
| SNPs | YCC Haplogroup - Long Form | YCC Haplogroup - Short Form |
|---|---|---|
| M230, P202, P204 | S | S-M230 |
| M254 | S1 | S-M254 |
| P57 | S1a | S-P57 |
| P61 | S1b | S-P61 |
| P83 | S1c | S-P83 |
| M226 | S1d | S-M226 |
Deep Clade Testing
Once you know your Y chromosome haplogroup, you may then focus on your branch of the tree through subclade testing. Testing begins with a predicted subclade. Enough SNPs are tested to identify and confirm your placement in the most current version of the YCC tree. Your results and placement on the tree are shown on the Haplotree.
Haplogroup E
Haplogroup E is one of the two branches of the mega-haplogroup DE. It originated approximately 50,000 years ago. Scientists believe that it ether arose in Africa or represents a back migration. It has been linked to the Neolithic expansion of peoples into Southern Europe. Over sixty subclades of E have been discovered.
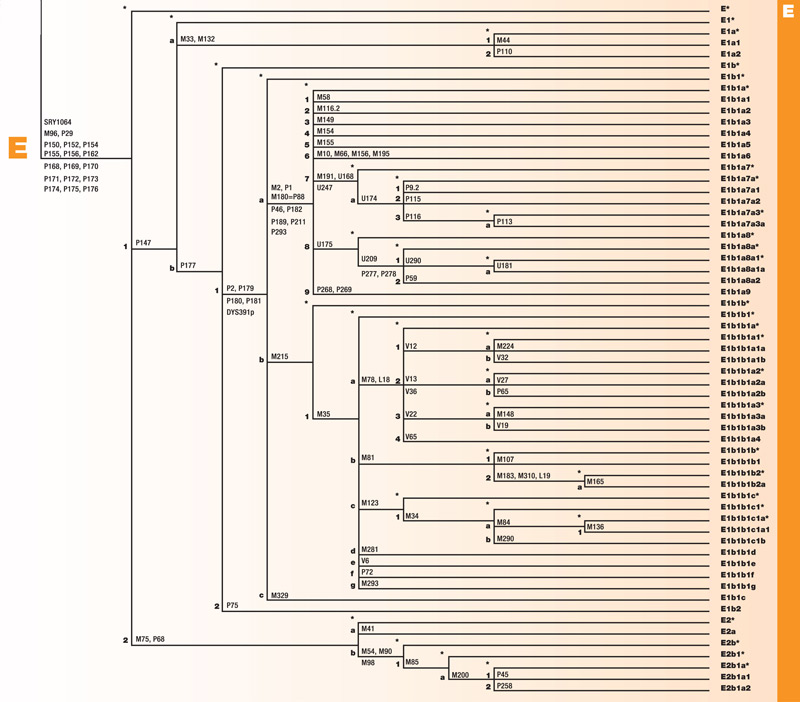
Haplogroup G
Haplogroup G is a branch of the mega-haplogroup F. G originated approximately 25,000 years ago in Eastern Africa. Its branches have spread into Eurasia. Some branches moved across Southern Asia and from there to India. Others moved across the Mediterranean and into Europe.
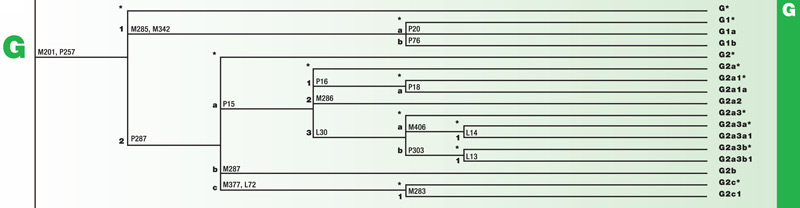
Haplogroup H
Haplogroup H is a branch of the mega-haplogroup F. H originated approximately 30,000 years ago in Eastern Africa. It spread to the Indian subcontinent and is found at high frequencies in India and Sri Lanka. It is also found in the Roma populations of Europe.

Haplogroup I
Haplogroup I is a branch of the mega-haplogroup F and its subsequent mega-haplogroup IJ. I originated approximately 25,000 years ago among the people of Eastern Africa and Southern Europe. As the ice receded after the last glacial maximum, it spread into Northern Europe.
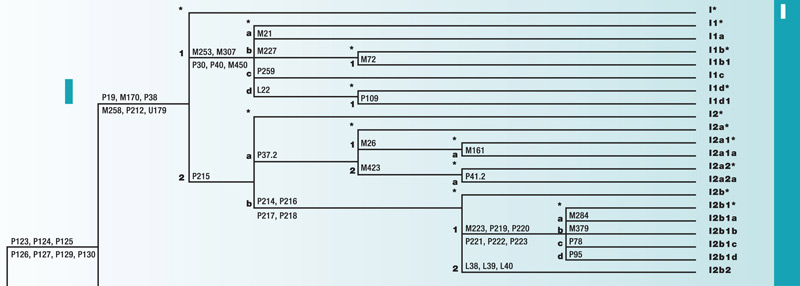
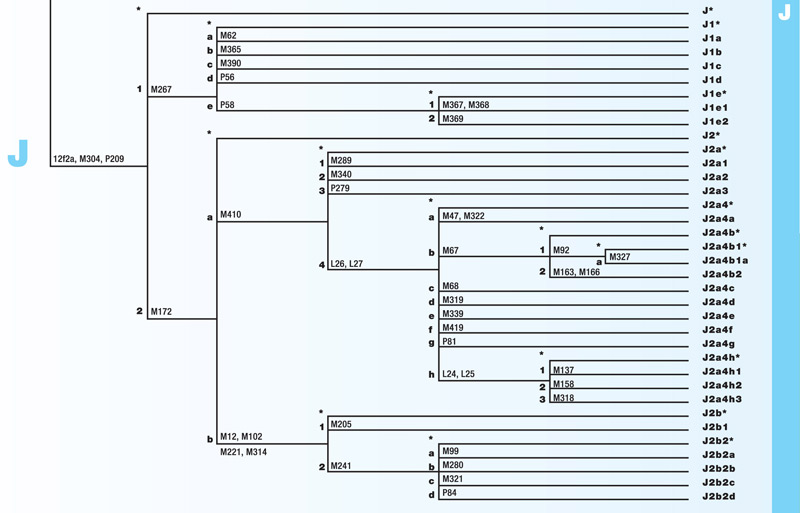
Haplogroup J
Haplogroup J is a branch of the mega-haplogroup F and its subsequent mega-haplogroup IJ. J originated approximately 25,000 years ago in the Eastern Africa Levant. It has two main branches, J1 and J2. Both are found in Eastern African populations. It has also spread into Europe and the Indian subcontinent during the Bronze Age. J1 is the parent haplogroup of the Cohen Model Haplotype, CMH.
Haplogroup N
Haplogroup N is a branch of the mega-haplogroup K. N originated approximately 10,000 years ago in Asia. Its branches have spread into East Asia and across Northern Europe.
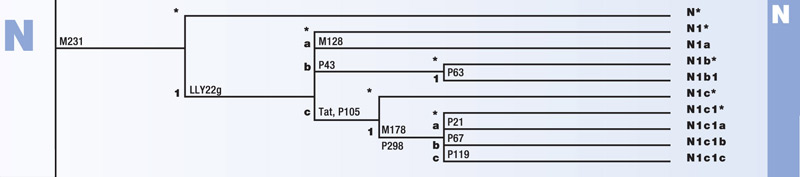
Haplogroup O
Haplogroup O is a branch of the mega-haplogroup K. O originated approximately 35,000 years ago in Asia. Its branches have spread into Central and East Asia. O has about thirty known subclades.
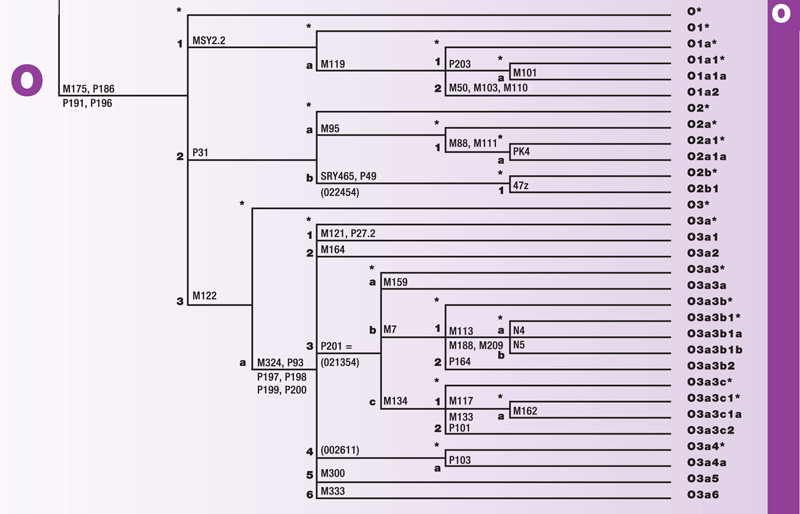
Haplogroup Q
Haplogroup Q is one of two branches of the mega-haplogroup P. Q originated approximately 20,000 years ago in Central Asia. Its branches have migrated into both Europe and East Asia. Some of its branches took part in the settlement of the Americas. These branches make up the majority of pre-Columbian Amerindian populations.
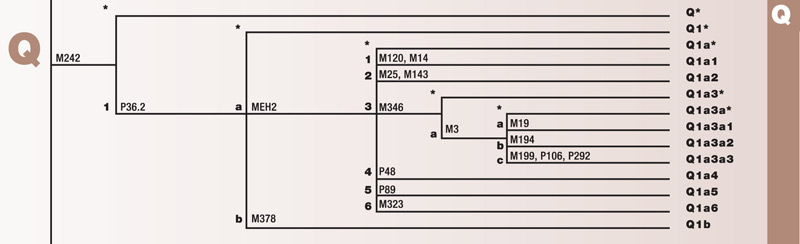
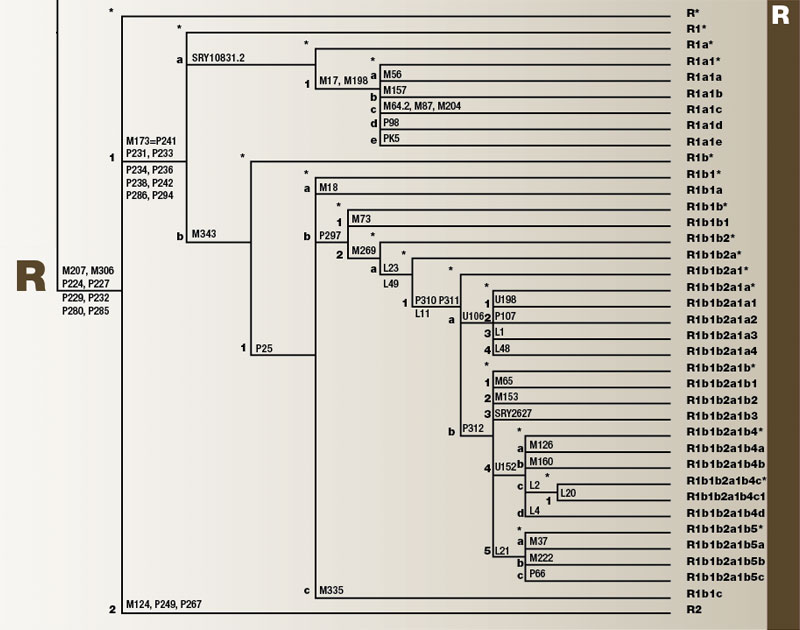
Haplogroup R
Haplogroup R is one of the two branches of the mega-haplogroup P. R originated approximately 30,000 years ago in Central Asia. It has two main branches, R1 and R2. R1 spread from Central Asia into Europe. Meanwhile, R2 spread east into the Indian subcontinent. Population movements have brought small numbers of both southward into the Eastern African Levant.
Matrilineal surname
Matrilineal surnames or mother-line surnames are inherited or handed down from mother to daughter (to daughter) in matrilineal cultures, similar to the more familiar patrilineal surnames which are inherited or handed down from father to son (to son) in patrilineal cultures (or societies).
For clarity and for brevity, the scientific terms patrilineal surname and matrilineal surname will usually be abbreviated as patriname and matriname, used interchangeably with fathername and mothername.
Mitochondrial DNA (mtDNA) testing
A person's matrilineal or mother-line ancestry can be traced using the DNA in his or her mitochondria, the mtDNA, as follows: This mtDNA is passed down by the mother unchanged, to all children. If a perfect match is found to another person's mtDNA test results, one may find a common ancestor in the other relative's (matrilineal) "information table", similar to the patrilineal or Y-DNA testing case above. However, because mtDNA mutations are very rare, a nearly perfect match is not as helpful as it is for the above patrilineal case. In the matrilineal case, it takes a perfect match to be very helpful.
Note that, in cultures lacking matrilineal surnames to pass down, neither relative above is likely to have as many generations of ancestors in their matrilineal information table as in the above patrilineal or Y-DNA case: for further information on this difficulty in traditional genealogy, due to lack of matrilineal surnames, see Matrilineality's section Matrilineal surname.
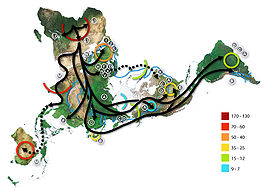
Map of human migration out of Africa, according to Mitochondrial DNA. The numbers represent thousands of years before present time. The blue line represents the area covered in ice or tundra during the last great ice age. The North Pole is at the center. Africa, the center of the start of the migration, is at the top left and South America is at the far right....
Some people cite paternal mtDNA transmission as invalidating mtDNA testing, but this has not been found problematic in genealogical DNA testing, nor in scholarly population genetics studies. See the rest of this article.
mtDNA by current conventions is divided into three regions. They are the coding region (00577-16023) and two Hyper Variable Regions (HVR1 [16024-16569], and HVR2 [00001-00576]). All test results are compared to the mtDNA of a European in Haplogroup H2a2a. This early sample is known as the Cambridge Reference Sequence (CRS). A list of single nucleotide polymorphisms (SNPs) is returned. The relatively few "mutations" or "transitions" that are found are then reported simply as differences from the CRS, such as in the examples just below.
The two most common mtDNA tests are a sequence of HVR1 and a sequence of both HVR1 and HVR2. Some mtDNA tests may only analyze a partial range in these regions. Some people are now choosing to have a full sequence performed, to maximize their genealogical help. The full sequence is still somewhat controversial because it may reveal medical information.
The most basic of mtDNA tests will sequence Hyper Variable Region 1 (HVR1). HVR1 nucleotides are numbered 16024-16569.[9 Some test reports might omit the 16 prefix from HVR1 results, i.e. 519C and not 16519C.

